Numerous studies examined the role of the microbiome in the onset of different pathologies, with most of them focusing on the intestinal microbiome.Recent studies have shown that alterations in the blood microbiota signature are associated with chronic inflammatory diseases, such as Diabetes, Obesity, and Cardiovascular diseases.This article briefly discussed blood microbiota profiles and cardiovascular disease.
Cardiovascular diseases (CVDs) are now the leading cause of morbidity and mortality worldwide (Khan et al., 2022).Despite substantial breakthroughs in prevention, fast detection, and treatment, CVDs continue to place a significant socioeconomic burden on health systems and communities. CVDs prevalence has risen by 93%, and mortality has risen by 54%, accounting for more than a third of all annual deaths globally(Roth et al., 2021). While non-communicable diseases account for 60% of disability-adjusted life-years worldwide. CVD accounts for roughly a quarter of this burden(Gheorghe et al., 2018).In addition to conventional risk factors, the role of recent changes in blood microbial mix in the progression of cardiovascular disease has gotten a lot of attention.
As human blood used to be considered a sterile environment composed of plasma, blood cells, and platelets, the presence of bacteria was considered a sign of infection (Katz, 1981).In the last two decades, multiple studies targeting the 16S ribosomal RNA gene have revealed the existence of a dominant phylum of blood-borne bacteria, which includes Proteobacteria, Actinobacteria, Firmicutes, and Bacteroidetes (McLaughlin et al., 2002; Amar et al., 2011, 2013; Dinakaran et al., 2014; Païssé et al., 2016; Gosiewski and Huminska, 2017; Loohuis et al., 2018; Qiu et al., 2019), Figure 1 shows that longer-term human blood microbiome constancy is indicated.In contrast, the microbiota structure in the blood rarely stays the same.
It has become increasingly popular to analyze the human microbiome because of the promise of gaining a greater understanding of biological systems.There has been previous work attempting to establish an evolutionary relationship between oral and gut bacteria, this study focuses on our blood microbial community.Developing a comprehensive understanding of the blood microbiome will undoubtedly lead to breakthroughs in how we treat diseases originating in the blood. “In addition to the therapeutic potential of tissue-specific microbiomes, this kind of analysis is highly sought after”. What is the significance of investigating the microbiome of the blood?.“Blood microbiomes research has important implications for environmental and medicinal microbiology. The environment is inextricably linked to our understanding of human microbiology and its surrounding bacteria”.Microbiology research includes not only the diagnosis, prognosis, and therapy of diseases but also the study of beneficial bacteria. As a result, microbiome analysis combats the prevention and treatment of numerous diseases.
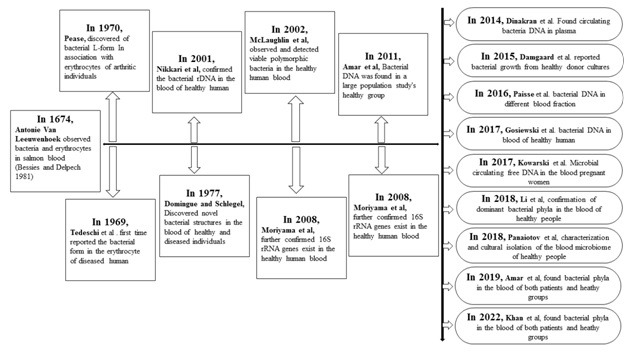
Figure. 1.Timeline indicating significant advances concerning human blood microbiome research
Microbiota in the blood of healthy individuals
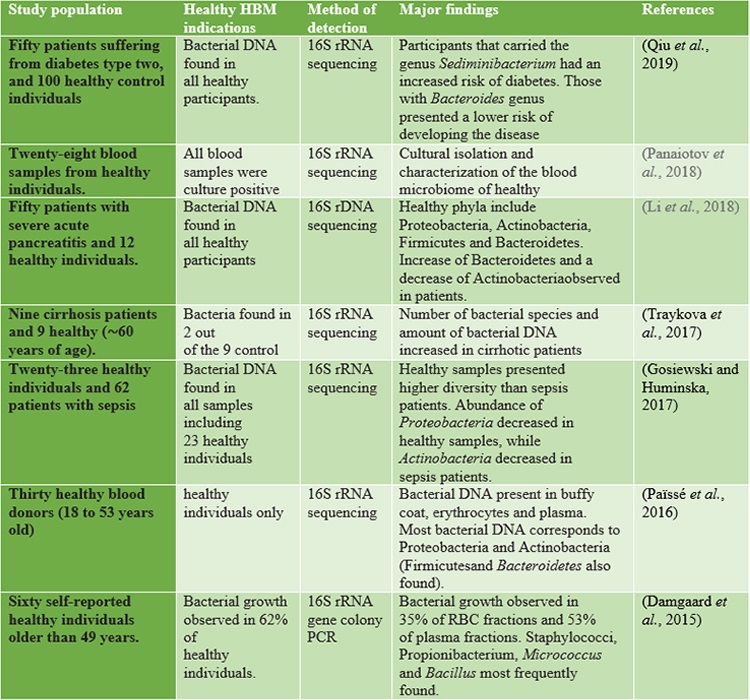
As already mentioned, 16S rRNA detection by sequencing is considered a hallmark of bacterial presenceand a way to assess microbial diversity since the advent of NGS-based tools(Tsamis et al., 2021). Recently, though,the attention of several groups has been drawn to the detection of 16S rRNA in “germ-free” niches,including the blood of healthy subjects (Ricci et al., 2020). As previous studies reported, the healthy human blood contained bacterial 16S rRNA, while the underlying mechanisms and its presence were not assessed. The only explanation involves that the bacterial transmission occurs from the mother before birth or translocation from other sites during the normal lifecycle(Nikkari et al., 2001). However, recent studies demonstrated that the blood microbiota in healthy humansis composed of the phylum Proteobacteria followed by Actinobacteria, Bacteroidetes, and Firmicutes, though alteration was observed across the differentstudies(Castillo et al., 2019). For instance, Paisse et al. investigated the blood microbiome composition in different fractions, they extracted DNA from red blood cells, buffy coats, and whole blood fractions of 30 healthy and young volunteers and performed quantitative PCR analysis and sequencing of V3-V4 hypervariableregions of the 16S rRNA for taxonomic classification. The highest abundance was observed in thebuffy coat (93.74% of bacterial DNA), followed by RBCs (6.23%) and plasma (0.03%), with the RBCfraction showing a higher bacterial diversity than the other two components (Paisse et al., 2016). Proteobacteria were found in more than (80%) of the blood and followed by Actinobacteria (6.7-10%) depending on the fraction, at variance from the phyla predominant in the gut (Firmicutes and Bacteroidetes)(Païssé et al., 2016). A healthy blood microbiome is given in Table 1.
Table.1. | Healthy blood microbiome studies concerning both healthy and diseased human participants.
Microbiota found in the blood of Cardiovascular diseases
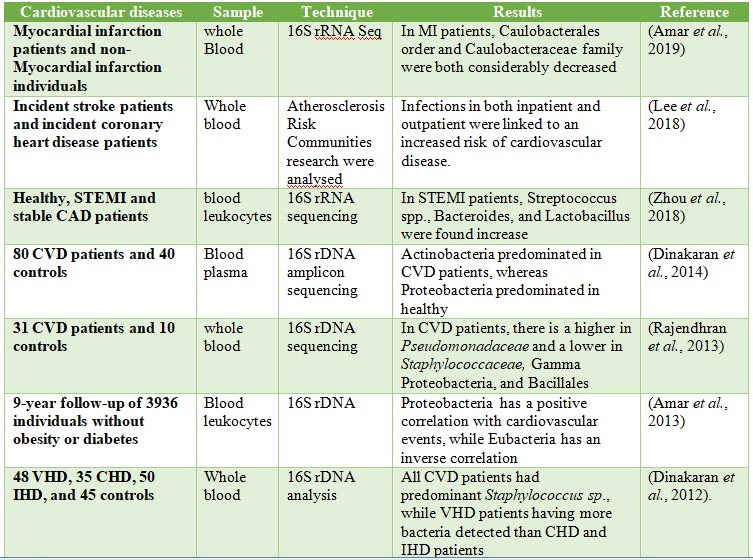
High-throughput sequencing and Culture-independent technologies can beused to assess the microbiome of each organ in the body athigh taxonomic resolution (Ross et al., 2019). In the year 2013, a study profiled microbial communities in whole blood of CVD patients and healthy controls by 16S rRNA sequencing(Rajendhran et al., 2013), though no variation in bacterial diversity at the phylum level was observed, an increase in Proteobacteria and a decrease in Firmicutes was reported. Using shotgun metagenome sequencing, the same group discovered a large increase in Pseudomonadaceae and decreases in Gammaproteobacteria, Bacillales, and Staphylococcaceae at lower taxonomic levels. Following that, it was discovered that CVD patients had a larger concentration of circulating plasma microbial DNA, as well as an increase in the Actinobacteria/Proteobacteria ratio (Dinakaran et al., 2012). Dinakaran et al. also reported that bacteriophages (Propionibacterium, Pseudomonas, and Rhizobium phages) predominate in the circulating virome of CVD patients, whereas eukaryotic viruses are predominant in healthy individuals (Dinakaran et al., 2014).
Additionally, Zhou et al. reported that microbial translocation from gut to blood is seen in ST-segment elevation myocardial infarction (STEMI) patients and mouse models as a result of disruption of the gut barrier composed of tight junction proteins. Increased detection of intestinal bacteria (Lactobacillus, Bacteroides, and Streptococcus) was observed in STEMI patients(Zhou et al. et al., 2018). More recently, Amar et al. reported a decreased abundance of cholesterol-degrading bacteria in the bloodmicrobiota of myocardial infarction patients, several (Norcardiaceae, Aerococcaceae, Gordonia, Propionibacterium, Chryseobacterium,Rhodococcus) are present in the blood of our control and case-patients, and remarkably the relative proportions of all ofthem are decreased in patients with MI. Moreover, the Caulobacterales order and the Caulobacteraceae family weresignificantly decreased in the MI group with both statisticalstrategies and their presence in theblood of patients with MI tended to be negatively correlatedwith LVEF at inclusion(Amar et al., 2019). We recently investigated the blood microbial composition in the Chines myocardial infarction patients and compare them with healthy individuals. We found that Actinobacteria were significantly higher in the blood of myocardial infarction patients (Khan et al., 2022). However, we did not observe the role of Actinobacteria in the onset of myocardial infarction. Further studies are worth needed to explore the role of blood microbiota in the onset of myocardial infarction. Various blood microbes are found in the blood of CVD patients, Tale 2.
Table 2. The microbiota was reported in the Blood of CVD Patients.
Issues to be addressed in the future
Most clinical studies compare the blood microbial composition between patients and healthy controls. And It is now recognized that blood is not a sterile environment, and several lines of evidence indicate that blood circulation is one of the major microbial niches of the human body. Moreover, recent studies have provided a useful characterization of the blood microbial profile in patients with CVD; however, we are still struggling with these descriptive data. A specific bloodmicrobiota-based target to prevent CVD has yet to emerge, which is the greatest challenge that weare currently facing.It may take a little more time to conduct a large cohort study or a translationalstudy to promote a deeper understanding of how the blood microbiota directly contributes to CVD. While Some researchershypothesized that the bacteria in the blood originated from gastro-intestinal tract leakage, it has alsobeen suggested that they could derive from skin or the oral tract and that they diffuse in blood whenthese protective barriers are compromised. However, it is important to note that the detection of microbial DNA/RNA in circulation cannot be directly correlated with the presence of live bacteria in the blood. Extensive changes in the blood microbiota and metabolome during CVD indicate that they may play a role in the etiology and development of the disease.Further research on understanding the molecular mechanisms of microbial translocation and their physiology in the development of disease will be necessary to establish the blood microbiota and their metabolites as biomarkers and therapeutic targets in CVD.
Conclusion
Thus, from the initial studies, it is evident that microbialcomponents exist in the circulation of CVDs which are normallyrecognized as non-infectious and non-communicable and these components might have a possible association with theclinical impact of CVDs. Blood microbiotain CVDs is dependent on patient characteristics like age, gender,glycemic status (diabetes mellitus), obesity, hypertension,smoking, alcohol consumption, and geneticpredisposition. Besides these factors, from the extensive researchduring the last decades, it is evident that diseases of the gut andoral cavity have a strong effect on the blood microbiome in CVDs. The development of the disease coulddetermine the microbiome diversity and load, thus elevating theinfectious burden which in turn, could exasperate the disease.
References
Amar, J., Lange, C., Payros, G., Garret, C., Chabo, C., Lantieri, O., et al. (2013). Blood microbiota dysbiosis is associated with the onset of cardiovascular events in a large general population: the DESIR study. PLoS One 8. doi:10.1371/journal.pone.0054461.
Amar, J., Lelouvier, B., Servant, F., Lluch, J., Burcelin, R., Bongard, V., et al. (2019). Blood Microbiota Modification After Myocardial Infarction Depends Upon Low-Density Lipoprotein Cholesterol Levels. J. Am. Heart Assoc. 8. doi:10.1161/JAHA.118.011797.
Amar, J., Serino, M., Lange, C., Chabo, C., Iacovoni, J., Mondot, S., et al. (2011). Involvement of tissue bacteria in the onset of diabetes in humans: evidence for a concept. Diabetologia 54, 3055–3061. doi:10.1007/s00125-011-2329-8.
Castillo, D. J., Rifkin, R. F., Cowan, D. A., and Potgieter, M. (2019). The healthy human blood microbiome: Fact or fiction? Front. Cell. Infect. Microbiol. 9, 1–12. doi:10.3389/fcimb.2019.00148.
Damgaard, C., Magnussen, K., Enevold, C., Nilsson, M., Tolker-Nielsen, T., Holmstrup, P., et al. (2015). Viable bacteria associated with red blood cells and plasma in freshly drawn blood donations. PLoS One 10, e0120826. doi:10.1371/journal.pone.0120826.
Dinakaran, V., John, L., Rathinavel, A., Gunasekaran, P., and Rajendhran, J. (2012). Prevalence of Bacteria in the Circulation of Cardiovascular Disease Patients, Madurai, India. Hear. Lung Circ. 21, 281–283. doi:10.1016/j.hlc.2012.02.007.
Dinakaran, V., Rathinavel, A., Pushpanathan, M., Sivakumar, R., Gunasekaran, P., and Rajendhran, J. (2014). Elevated levels of circulating DNA in cardiovascular disease patients: Metagenomic profiling of microbiome in the circulation. PLoS One 9. doi:10.1371/journal.pone.0105221.
Gheorghe, A., Griffiths, U., Murphy, A., Legido-Quigley, H., Lamptey, P., and Perel, P. (2018). The economic burden of cardiovascular disease and hypertension in low-and middle-income countries: a systematic.
Gosiewski, T., and Huminska, K. (2017). Comprehensive detection and identification of bacterial DNA in the blood of patients with sepsis and healthy volunteers using next-generation sequencing method - the observation of DNAemia. Eur. J. Clin. Microbiol. Infect. Dis., 329–336. doi:10.1007/s10096-016-2805-7.
Katz, P. (1981). Ritual in the operating room. Ethnology 20, 335–350.
Khan, I., Khan, I., Jianye, Z., Xiaohua, Z., Khan, M., Gul, M., et al. (2022). Exploring blood microbial communities and their influence on human cardiovascular disease. 1–11. doi:10.1002/jcla.24354.
Lee, H. J., Seo, H. I., Cha, H. Y., Yang, Y. J., Kwon, S. H., and Yang, S. J. (2018). Diabetes and Alzheimer’s Disease: Mechanisms and Nutritional Aspects. Clin. Nutr. Res. 7, 229. doi:10.7762/cnr.2018.7.4.229.
Li, Q., Wang, C., Tang, C., Zhao, X., He, Q., and Li, J. (2018). Identification and characterization of blood and neutrophil-associated microbiomes in patients with severe acute pancreatitis using next-generation sequencing. Front. Cell. Infect. Microbiol. 8, 1–16. doi:10.3389/fcimb.2018.00005.
Loohuis, L., Mangul, S., Ori, A., … G. J.-T., and 2018, U. (2018). Transcriptome analysis in whole blood reveals increased microbial diversity in schizophrenia. nature.com. doi:10.1038/s41398-018-0107-9.
McLaughlin, R. W., Vali, H., Lau, P. C. K. K., Palfree, R. G. E. E., De Ciccio, A., Sirois, M., et al. (2002). Are there naturally occurring pleomorphic bacteria in the blood of healthy humans? J. Clin. Microbiol. 40, 4771–4775. doi:10.1128/JCM.40.12.4771-4775.2002.
Nikkari, S., McLaughlin, I. J., Bi, W., Dodge, D. E., and Relman, D. A. (2001). Does the blood of healthy subjects contain bacterial ribosomal DNA? J. Clin. Microbiol. 39, 1956–1959. doi:10.1128/JCM.39.5.1956-1959.2001.
Païssé, S., Valle, C., Servant, F., Courtney, M., Burcelin, R., Amar, J., et al. (2016). A comprehensive description of blood microbiome from healthy donors assessed by 16S targeted metagenomic sequencing. Transfusion 56, 1138–1147. doi:10.1111/trf.13477.
Panaiotov, S., Filevski, G., Equestre, M., Nikolova, E., and Kalfin, R. (2018). Cultural isolation and characteristics of the blood microbiome of healthy individuals. Adv. Microbiol. 8, 406–421. doi:10.4236/aim.2018.85027.
Qiu, J., Zhou, H., Jing, Y., Dong, C., Laboratory, C. D.-J. of clinical, and 2019, U. (2019). Association between blood microbiome and type 2 diabetes mellitus: A nested case‐control study. J. Clin. Lab. Anal. 33, e22842. doi:10.1002/jcla.22842.
Rajendhran, J., Shankar, M., Dinakaran, V., Rathinavel, A., and Gunasekaran, P. (2013). Contrasting circulating microbiome in cardiovascular disease patients and healthy individuals. Int. J. Cardiol. 168, 5118–5120. doi:10.1016/j.ijcard.2013.07.232.
Ricci, V., Carcione, D., Messina, S., Colombo, G. I., D’alessandra, Y., and D’alessandra, Y. (2020). Circulating 16s rna in biofluids: Extracellular vesicles as mirrors of human microbiome? Int. J. Mol. Sci. 21, 1–14. doi:10.3390/ijms21238959.
Ross, A. A., Rodrigues Hoffmann, A., and Neufeld, J. D. (2019). The skin microbiome of vertebrates. Microbiome 7, 1–14.
Roth, G. A., Mensah, G. A., Johnson, C. O., Addolorato, G., Ammirati, E., Baddour, L. M., et al. (2021). Global Burden of Cardiovascular Diseases and Risk Factors, 1990-2019: Update From the GBD 2019 Study (vol 76, pg 2982, 2020). J. Am. Coll. Cardiol. 77, 1958–1959.
Traykova, D., Schneider, B., Chojkier, M., and Buck, M. (2017). Blood microbiome quantity and the hyperdynamic circulation in decompensated cirrhotic patients. PLoS One 12, e0169310.
Tsamis, K. I., Sakkas, H., Giannakis, A., Ryu, H. S., Gartzonika, C., and Nikas, I. P. (2021). Evaluating Infectious, Neoplastic, Immunological, and Degenerative Diseases of the Central Nervous System with Cerebrospinal fluid-based Next-Generation Sequencing. Mol. Diagn. Ther. doi:10.1007/s40291-021-00513-x.
Zhou et al. (2018). Gut-dependent microbial translocation induces inflammation and cardiovascular events after ST-elevation myocardial infarction. Microbiome 6, 1–17. doi:10.1186/s40168-018-0441-4.