The article focuses on the use of MALDI-TOF mass spectrometry (MS) for advancing clinical microbial identification, specifically in the Microbiology Laboratory at the Department of Laboratory Medicine, Beijing Tongren Hospital, which has sought to find an alternative to traditional culturing methods for pathogen diagnosis.The article also explains the benefits MALDITOF MS technology has had on the lab and why conventional methods are not suitable for identification of challenging microorganisms, such as those causing fungal rhinosinusitis (FRS).
Clinical microbiology research plays a vital role in improving the diagnosis of infectious diseases, drug guidance, hospital infection control and antimicrobial drug management. Traditional biochemical identification methods are relatively complex and cumbersome and cannot fully meet the requirements of turnover time, sample diversity, and identification accuracy. As a result, hospital laboratories are seeking alternatives to traditional culturing methods for pathogen diagnosis.
Unlike traditional biochemical methods, matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry (MALDI-TOF MS) determines the protein profile of microbial highabundance proteins (mostly ribosome proteins) and compares their characteristic spectra with proprietary libraries holding reference spectra. Most microorganisms can be accurately identified at the species level. Species-conserved protein sequencing, extensive library references, and strict library quality control processes ensure the accuracy of MALDI-TOF MS identification.
Hospital-wide benefits
Using advanced MALDI-TOF MS technology, a single sample preparation can take only 20 seconds, and approximately 100 samples can be analysed in 60 minutes, greatly improving the speed of identification and the through put ofthe hospital laboratory. For critically ill patients, obtaining the results 24 hours sooner than with traditional methods is important for rapid targeted antibiotic treatment.
The financial advantages of MALDITOF MS are also significantly changing clinical workflows, as the cost of biochemical identification typically prohibits the analysis of every potentially contaminating bacterium, meaning some significant microbes may be missed. MS breaks down this barrier, with reagent costs nearly 10 times lower than that of biochemical methods. Identification accuracy is another key benefit. The difficult pathogens that cannot be identified using routine biochemical approaches, such as anaerobic bacteria, can be accurately identified using MALDI-TOF MS. The MS database covers all types of bacteria and fungi, with a clinically common pathogen identification accuracy of more than 98 per cent.
Identifying challenging fungi
Unlike common bacteria and yeast, challenging pathogens are difficult to culture, and have slow metabolisms, diverse growth periods, and complex cell wall structures, making them hard to identify by conventional methods. Filamentous fungi are a key example of pathogens that have, until recently, required cumbersome identification methods involving cultures, microscopic examination, or molecular sequencing. In addition, the identification is highly dependent on the technical capability and experience of laboratory technicians. Fungal rhinosinusitis (FRS) – a broad term referring to a group of conditions caused by fungal infections of the paranasal sinuses – can cause serious disease if not treated promptly and with appropriate anti-fungal drugs. Misuse of anti-fungals and environmental pollution are both significant contributors to the severity of FRS.
Misdiagnosis of FRS, particularly in the case of acute invasive FRS, may lead to treatment failure and can be fatal. Studies show that 80 per cent of FRS is caused by common pathogenic fungi Aspergillus spp. In addition, Fusarium spp., Mucor spp., Rhizopus spp., Rhizomucor spp., Penicillium spp. and Alternaria spp. could be causative agents. Conventional FRS diagnostic methods include clinical observation, endoscopic examination, imaging, pathology, and fungal morphology identification.
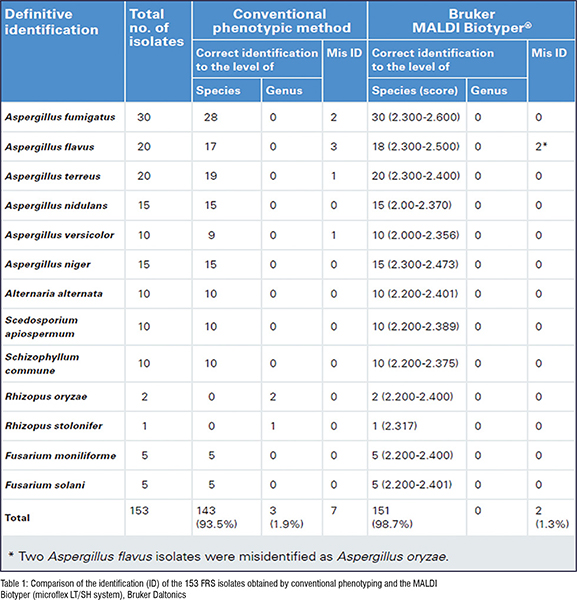
FRS pathogens are particularly challenging to identify in clinical laboratories, due to the wide variety of causative species with phenotypically similar characteristics, despite their genetic differences. Identification relies heavily on the experience of laboratory technicians. Identifying FRS pathogens with conventional methods often incurs high costs, results in long turnaround times, low throughput, and low selectivity, and cannot exclude colonisation. Conventional methods are therefore not suitable for the identification of FRS pathogens. MALDI-TOF MS can identify the individual species using direct transfer to the target or extraction methods, and shortens the identification time of fungi species, bringing new possibilities for understanding and treating FRS.
In 2017, the Microbiology Laboratory of Beijing Tongren Hospital evaluated the performance of MALDI-TOF MS (microflex LT/SH system, Bruker Daltonics, Bremen) in identifying 153 different FRS filamentous fungi isolates.1 The results showed that 151 out of 153 FRS isolates had correct species identification (98.7 per cent) (Table 1). The rate was significantly higher than that of the traditional phenotypic methods (93.5 per cent). MALDI-TOF MS was capable of identifying very closely related species including Aspergillus versicolor (1), Aspergillus flavus (3), Aspergillus fumigatus (2) and Aspergillus terreus (1), which were indistinguishable with conventional phenotypic methods. The MALDI-TOF MS method has proven to be more accurate and quicker than phenotypic methods for the identification of filamentous fungi in clinical microbiology laboratories.
Tacking antimicrobial resistance
MS-based microbial methods are not only changing the way clinical pathogens are identified, but are also aiding the fight against antimicrobial resistance. Carbapenem-resistant Enterobacteriaceae (CRE) are a group of bacteria that are resistant to the last-line antibiotic carbapenem and pose a significant public health threat as a multi-drug resistant (MDR) pathogen. A study published in 2013 shows the ability of MALDITOF MS to detect carbapenem resistance in VIM-2-producing Pseudomonas aeruginosa, AmpC- and KPC-producing carbapenem resistant Enterobacteriaceae, and OXA-23-producing Acinetobacter baumannii.2 The detection accuracy rate of MALDI-TOF MS for enzyme-producing and non-enzyme-producing strains is up to 100 per cent, which proves that MALDI-TOF MS has great advantages over other existing phenotypic methods on carbapenemase detection.
The national average resistance rate for carbapenem-resistant Klebsiella pneumonia in China has reached 10.1 per cent (according to 2018 National Bacterial Resistance Detection Network)3 since it was first reported in Zhejiang Province in China in 2007. The mortality of nosocomial infections caused by CRE is as high as 33.5 per cent, and bloodstream infections are as high as 43.1 per cent.4 Because it is easy to spread, enzyme-producing strains accounted for more than 90 per cent of CRE. Rapid detection of enzyme-producing strains is very important for timely and effective clinical treatment. It is also beneficial to promote the rational application of antibiotics and ease the severe situation of antibiotic resistance.
Looking forward
As an emerging rapid detection technology, MALDI-TOF MS is gaining traction as a routine clinical identification tool. The technology has undergone rapid development in clinical microbiological testing in China over the past ten years, and further advancements such as bacterial typing and homologues analysis, pathogen antigens, serum antibodies, toxicity detection and pathogen identification through fluorescence, could make MALDI-TOF MS a complete solution for microbiologists over the next decade.