Pharma has begun incorporating real-world data (RWD) and real-world evidence (RWE) into the process of clinical development, primarily to facilitate clinical trials and preparation for submission to regulatory agencies, e.g. FDA, EMEA. In a previous article, we pointed out the opportunity to enhance product value and now can show how this can be accomplished using novel analytics applied to real world data in both drug discovery and development
Drug development, today, is a risky and expensive business. Drug discovery and development exhibits a 90 per cent failure rate that as a process can take between 10-15 years and whose average cost is US$1-2 billion/newly approved drug. The appearance of toxic side effects and/or the lack of efficacy highlights the fact that human patients are different and more complex than the animal and cell models used in early development. Even for drugs that achieve regulatory approval, commercial success is not guaranteed, financially affecting both pharma and payers (public and private). Among the diverse challenges in this complex process are those that involve technology, science, regulatory oversight, financial issues, and the sociology, culture and psychology of both the physician and the patient. Currently, significant efforts are underway to evaluate and incorporate the use of artificial intelligence and real world data/real world evidence to enhance the probability for success. One critical consideration is whether these approaches are actually attacking the root cause problems or are being constrained by ‘pipeline vision’, i.e. the need to continue to support the current drug development pipeline model.
Pharma, over many years and across the industry, has evolved and operates common drug discovery and development ‘pipelines’ that influence internal organisation and infrastructure. In general, pharma adopts new technology in an effort to optimise performance while only. slowly evolving its infrastructure and culture to support it. Optimisation can be viewed as ‘attempting to improve efficiency (and effectiveness?) of the pipeline model by reducing the ‘entropy at specific points along the path, e.g. target selection, target validation, drug discovery, clinical trial design, patient recruitment, regulatory submission, etc. In addition, the FDA’s does not require understanding the mechanism of action for a drug that is submitted for regulatory approval. As a result, drug discovery can add ‘phenotypic discovery and validation’. to its traditional ‘target selection and validation’. Below, in this article, the opportunity for real world data to redefine ‘phenotype’ and improve target selection will be discussed.
The inefficiencies in the current process are not uniformly distributed, ranging from 3 per cent (target validation) to 6 per cent (compound screening) and 66.4 per cent (Phase I), 48.6 per cent (Phase II) and 59 per cent (Phase III), respectively. A recent analysis of the average cost for each stage of drug discovery and development and final cost suggests that: ~$1B drug discovery and lead optimisation,
>$300M on preclinical studies and
> $50M (Phase I clinical trials)
>$100M (Phase II clinical trials)
>$300M (Phase III clinical trials).
< $10M regulatory review and approval
---------------------------------------------
$1.7B average total cost (13.5 years and increased from $1.5B in 2018)
Pharma has adopted the concept of a ‘pipeline’, borrowed from the petroleum industry, to describe the linear alignment of steps in drug discovery and development, through clinical trials and regulatory submission/approval. While this provides a convenient visualisation, i.e. of a linear process, there are additional characteristics of real pipelines whose considerations in drug development could further benefit drug development beyond a visual model. A true pipeline has pumps, valves and control devices and is subject to leaks, blockages and contamination. These elements can also be mapped to drug development and can provide additional critical insights.
This article focuses on the issues of pipeline leaks and contamination, i.e. development of potentially good drugs but addressing the wrong target and inadequate understanding of the complexity of the patient, of the disease and of the practice of medicine. Some re-direction of the use of real-world data could contribute to closing these gaps.
Currently, two somewhat divergent approaches to improve successful drug development have been adopted in large pharma: 1) internal investment in access and application of new technologies that result from exciting, new scientific breakthroughs, and 2) outsourcing/licensing/ investing involving small biotechnology and technology companies for early access to potential products to minimise dependence on the less efficient parts of the current discovery process.
-A potential challenge to implementing the first approach is that most new technologies are applied to the existing pipeline model rather than exploring whether ‘pipeline redesign’ might provide a better solution, because of ‘pipeline vision’. Re-engineering the pharma pipeline would require significant disruption to both its existing infrastructure but even more, its culture, i.e. its people. Improving efficiency is a valid target, but this focuses on speed, i.e. ‘fail fast’, and may not address the ‘leaks and contamination’ in the pipeline, discussed later in this article, which are lessons to be learned for further drug development.
-In the second approach, outsourcing is effectively carried out by modest investment, e.g.
The recognition of the potential value currently encapsulated in rich, real world data sources has evolved along with advances in artificial intelligence and machine learning that can manage large data sets in an automated manner to identify patterns that might be difficult for an individual researcher. The emerging field of Big Data analytics, using the aggregate of real world data as noted above, suggests potential benefits may include:
Even this small subset of all potential uses of Big Data and analytics could provide great value to pharma/biotech industry, physician decision making and, most importantly, the patient, but even greater value might be recognised by addressing the challenges that remain. I am reminded of this quote (actually acknowledged by Laurie Anderson, 2020, as being borrowed)
"If you think technology will solve your problems,
— you don’t understand technology
— and you don’t understand your
problems."
I would rephrase the last line to “you don’t fully understand the complexity of the problem and its underlying challenges”.
The poor success rate in drug development may be the impact of not addressing or even acknowledging these challenges without recognising that this may result from ‘pipeline vision’. With an infrastructure and culture that focuses on supporting and implementing the drug pipeline model, new technologies and new data are being applied to the existing model rather than exploring whether they are addressing critical, underlying, and complex questions along the pipeline. The alignment of computational and informatic technologies along the pipeline are shown in Figure 1 which further delineates the pipeline in terms of three stages: guided exploration, controlled validation and real-world validation, where validation includes Verification. Currently the primary use of real-world data is in these latter two stages; the opportunity is to use it to address the first stage, hence enable ‘fail faster’ and increase the potential for greater success in drug discovery, regulatory approval and clinical acceptance.
Drug Discovery typically begins with the biological identification and assessment of a target molecule, e.g. protein, receptor, RNA, DNA, etc., and progresses to either small molecule or biologic selection or development that exhibits selectivity and specificity for that target. The ideal target would represent an early step along the known mechanism of the disease but this is not typically well understood. Currently RWD/ RWE contributes indirectly to this by evaluating which drugs have been more effective or exhibit reduced side-effects in real world populations so that potentially relevant pathways and additional molecular targets can be identified. This approach is further supported by use of machine learning methods to analyse RWD/RWE.
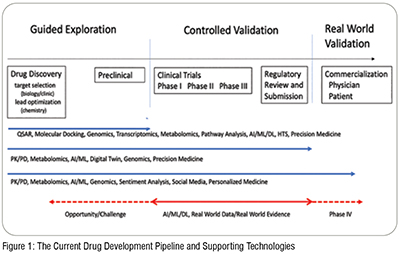
The drug discovery component of drug development consists of two phases: the biology phase and the chemistry phase. The biology phase precedes the chemistry phase as it focuses on the identification and qualification of the target, i.e. biologic process and molecular entity, through biological (and clinical) analysis. While this requires a comprehensive understanding of the disease process, ideally it should also includeunderstanding the complexity of the realworld patient and also guidelines and patterns of diagnosis and treatment. This is where RWD/RWE can contribute most significantly. Many of the computational approaches shown in Figure 1 (and also experimental approaches) involve chemical analysis and are applied to refine the lead compound, its physical, chemical and biochemical properties, and its potential selectivity and specificity for a target, i.e. lead optimisation.
Although seemingly obvious, the most critical phase of target identification (or phenotype selection) involves the accurate definition of the disease, itself. Many (most) diseases actually represent complex sets of disorders that are grouped within broad diagnostic categories, e.g. Migraine. Failure to recognise the critical differences among ‘disease subtypes’ can lead to drug-target success through clinical trials where controlled selection of patients occurs. Much more costly, however, is failure in the clinic or a drug that only addresses a small segment of the real world patient population and may not lead to a commercially viable product. It is common that clinical trials utilise inclusion/exclusion criteria that do not accurately reflect the real-world patient population, e.g. exclusion of women or lack of diversity, because their goal is to establish efficacy and safety. The drug discovery step presents the optimal opportunity to more broadly analyse (and understand) the real-world complexity of the disease, of the intended patient and even of the practice of medicine so that the drugs being developed can have a higher rate of success going from discovery to validation to regulatory approval and commercialisation, i.e. physician and patient acceptance and adherence. Real-world data and evidence, when appropriately aggregated and analysed, can significantly enhance the probability for success and provide additional verification. This first requires re-examining the definitions/usage of ‘disease’ and ‘phenotype’. Phenotype is commonly defined as the expression of one’s genomic makeup under the influence of environmental factors.
It is critical to recognise and incorporate the reality that disease is a process and not a state. This means that disease progresses over time and actually in a high-dimensional space that includes both clinical, e.g. lab results, and non-clinical, e.g. diet, environment, lifestyle, factors. That very over time. Access to much of this data does not exist or is variable in quality, and
Which factors are most relevant for any given disease is also unknown. Ideally we should consider 3 key elements to diagnose disease more accurately than we do: 1) disease trajectory, what is the high-dimensional vector that defines how the patient is progressing over time; 2) disease staging, how far along that vector is the patient when presenting for diagnosis; and, 3) disease velocity, how rapidly is the patient progressing. The result of not having such ideal data is that 1) a patient coming in for diagnosis at different stages of the disease may be diagnosed differently, 2) two patients may appear to be identical in terms of lab results but actually have different diseases (and progressions), 3) two patients may appear different in their labs but have the same disease, just presented for diagnosis at different stages of the disease. The further reality is that an average patient has 5 co-morbid conditions and these may be previously diagnosed and treated, currently diagnosed and being treated, undiagnosed and as yet untreated. These co-morbidities can significantly impact the disease trajectory and resulting diagnosis, treatment decision and response. These realities all present challenges, i.e. ‘leaks’ that impact our ‘pipeline vision’.
Phenotype is commonly defined as the expression of one’s genomic makeup under the influence of environmental factors. The concept of ‘environmental influence’ needs to consider factors beyond the conventional definition of environment. As noted above, co-morbidities, poly-pharmacy, lifestyle, social determinants of health and cultural determinants are examples of ‘environmental factors:’ that contribute to how genomic factors may or may not be expressed in an individual, and these factors may change over time. It is critical to re-examine current definitions of phenotype from seeking common observable factors in patients with the same diagnosis, to actually use the changes in these factors over time, which include clinical measurements, to define the ‘next generation phenotype’ and establish datadriven diagnoses of disease sub-types.
The lack of requirement for understanding mechanism of action for FDA approval, focusing on safety and efficacy, reinforces the use of correlative approaches for drug discovery and development rather than addressing the difficult study of causality. Many of the AI/ML methods currently used in early drug discovery further support these approaches utilising the increasing access to big data.
Real world data can contribute significantly to supporting the critical evolution from correlation to causality and approaching better definitions of disease and mechanisms of action but there remain challenges. To understand how real-world data and evidence might be used to ‘seal leaks in the pipeline’ and enhance the efficiency and the effectiveness of drug development, the common sources of the data must be considered. In a recent industry survey where more than 70 per cent were strongly committed to its use already, it was noted that more than half of the organisations surveyed used disease and product registries and electronic health records, with patient surveys, insurance claims, pharmacy records, digital health/monitoring/ wearables and imaging as data sources. Interesting was the observation that genomics data trailed these in terms of its use. It is also important to remember that a leak can impact a pipeline in at least two different ways: to lose materials, i.e. data, that results in inefficiency; and can contaminate the material, i.e. data, that remains within the pipeline because it is not aligned with what is needed. The application of AI/ML methods represents an opportunity for analysing large amounts of data and identifying critical patterns difficult to visualise, but the thirst for big data to support this needs to be cognisant of what the data actually does and does not represent.
As shown in Figure 1, RWD comprises at least two major sub-groups, business data (BD) and clinical data (CD). Each may be valid and useful but are developed to address different needs and this must be considered when using, and especially combining, them for analysis purposes. Is the goal to address the disease process at the molecular and clinical level or is it to examine associated business practices?
-Perhaps the most abundant data source in healthcare is claims data. This is acknowledged by physicians to not adequately represent the patient and their disease but rather what must be documented to support diagnostic testing, treatment procedures and drugs. In addition, it must bear some alignment with standard of care and clinical guidelines. As such, it is not a reliable source of data to define the true disease course, i.e. next generation phenotype and disease subtype, particularly as recent studies have shown that, on average, only just over 50 per cent of the time and their patients compliance with physician recommendations only achieves 54 per cent.
-this extends to the use of ICD-10 codes to define disease and disease progression as well -even reliance on EHR data to define the patient journey can be inadequate, particularly when combining records, i.e. the issue of interoperability of patient data. Many data scientists rely on matching of data fields and use of machine learning methods to assist in this process, to further support the development of big data sets for analysis. It is critical to understand that many of the data fields do not reflect the underlying complexity of the data entered, whose numerical value may fit well into use in algorithmic approaches. For example, blood pressure measurements are recorded but without definition of the method used to measure them, whether a patient had been resting for a period prior to measurement or just ‘run up the stairs to their appointment’, etc. It is important to understand that data quality needs to be examined and prioritised over data quantity even when using clinical records.
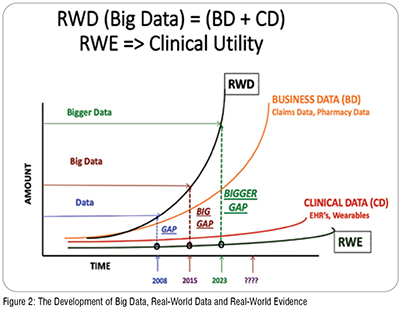
The increased access to wearable data and to patient records, to both claims data and to clinical (EHR) data, continues to expand the content of RWD but does not guarantee its conversion to RWE, particularly in light of the need to redefine the concept of phenotype and disease stratification to support and enhance both target selection and phenotypic screening for drug development remains a challenge. Figure 2.
While the development of drugs remains primarily a commercial activity, even if initiation may take place in an academic research laboratory, the ultimate beneficiary will always be the patient with the physician serving as the intermediary. Several studies point to significant errors in diagnosis, averaging 5 per cent (12 million patients) for all outpatients and 20 per cent for those with severe medical conditions and resulting in 40,000-80,000 deaths. In addition, the FDA reports that more than 100,000 medication errors are reported annually.
Addressing many of the issues raised above could serve to enhance the cooperation and collaboration between the physician, who deals with the real-world patient, and pharma to benefit all. The challenge is to recognise the truth in Anderson’s quote. Technology, alone, will not solve the complex problems in drug development and healthcare. It is critical to re-evaluate the perspectives that have evolved and expand drug discovery and development beyond the constraints of ‘pipeline vision’. This would seem to be scientifically, economically and humanitarily needed and valued. It is not likely that the technology will produce solutions to real-world problems if we do not first take the time to acknowledge the complexity of the problems, themselves. While it may seem simple to keep ‘rolling the rock up the hill’ as Sisyphus came to learn, ‘complexity keeps it from reaching the top’.